MENÚ
ES | EUR
ES | EUR
-
- Todas las centrífugas
- Centrífugas de sobremesa
- Centrífugas de suelo
- Centrífugas refrigeradas
- Microcentrífugas
- Centrífugas multiuso
- Centrífugas de alta velocidad
- Ultracentrífugas
- Concentrador
- Productos IVD
- High-Speed and Ultracentrifuge Consumables
- Tubos de centrífuga
- Placas de centrífuga
- Gestión de dispositivos
- Gestión de muestras e información
-
- Todas las pipetas, dispensadores y manipuladores automatizados de líquidos
- Pipetas mecánicas
- Pipetas electrónicas
- Pipetas multicanal
- Pipetas de desplazamiento positivo y dispensadores
- Puntas de pipeta
- Dispensadores de botella
- Controladores de pipeta
- Accesorios para dispensadores y pipetas
- Pipeteo automatizado
- Consumibles de automatización
- Accesorios de automatización
- Servicios para dispensadores y pipetas
Sorry, we couldn't find anything on our website containing your search term.
Sorry, we couldn't find anything on our website containing your search term.
How to identify eukaryotic contamination in your cell culture
Academia de laboratorio
- Biología celular
- Cultivo celular
- Contaminación
- Consumibles para cultivo celular
- Incubadores de CO2
- Ensayo
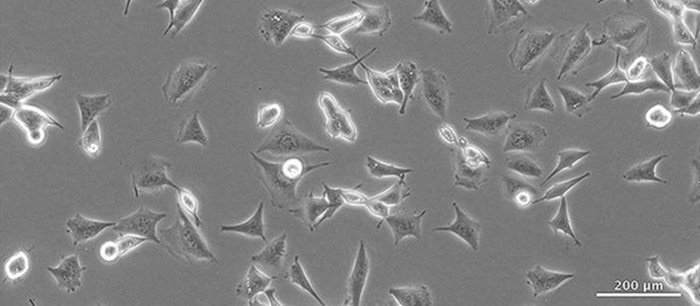
Eukaryotic contamination means that one cell line is contaminated with another cell line. There is no visible sign of a mixed culture.
The ICLAC (International cell line authentication committee) reports roughly 500 documented eukaryotic cell lines that are cross-contaminated with cells from other cell lines or misidentified. Current estimates assume that at least 15 % of all cell lines in use worldwide are cross-contaminated or identified as the wrong cell line. For these reasons, Good Cell Culture Practice documentation is paramount; not only to protect cells from contaminations such as bacteria and fungi, but also from each other. Misidentified and cross-contaminated cell lines can be detected by authentication testing. Short tandem repeat (STR) profiling is an accepted consensus method for authenticating human cell lines. Source: www.iclac.org
Find more about misidentified cell lines here .
Macroscopic detection
There are no means by which cross contamination can be detected macroscopically. Pay close attention if the growth rate of your cell line begins to differ from previous measurements/observations.Microscopic detection
Even with the help of a microscope it is difficult to detect cross contaminations with other eukaryotic cells. Sometimes the shape or dimension of the cell could provide a hint.The ICLAC (International cell line authentication committee) reports roughly 500 documented eukaryotic cell lines that are cross-contaminated with cells from other cell lines or misidentified. Current estimates assume that at least 15 % of all cell lines in use worldwide are cross-contaminated or identified as the wrong cell line. For these reasons, Good Cell Culture Practice documentation is paramount; not only to protect cells from contaminations such as bacteria and fungi, but also from each other. Misidentified and cross-contaminated cell lines can be detected by authentication testing. Short tandem repeat (STR) profiling is an accepted consensus method for authenticating human cell lines. Source: www.iclac.org
Find more about misidentified cell lines here .
Leer más
Related documents


PDF 2,10 MB
Related links
Leer más
